RNA
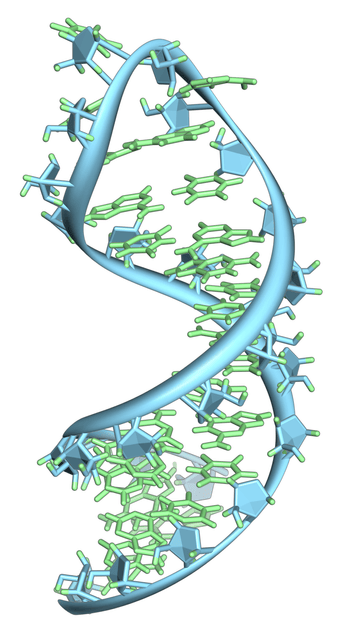
RNA

Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and DNA are nucleic acids, and, along with lipids, proteins and carbohydrates, constitute the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA it is more often found in nature as a single-strand folded onto itself, rather than a paired double-strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information (using the nitrogenous bases of guanine, uracil, adenine, and cytosine, denoted by the letters G, U, A, and C) that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.
Some RNA molecules play an active role within cells by catalyzing biological reactions, controlling gene expression, or sensing and communicating responses to cellular signals. One of these active processes is protein synthesis, a universal function in which RNA molecules direct the synthesis of proteins on ribosomes. This process uses transfer RNA (tRNA) molecules to deliver amino acids to the ribosome, where ribosomal RNA (rRNA) then links amino acids together to form coded proteins.
Comparison with DNA
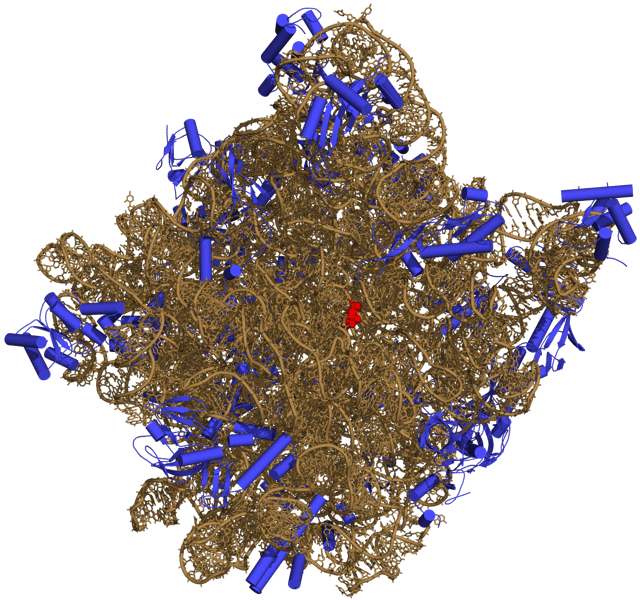
Three-dimensional representation of the 50S ribosomal subunit. Ribosomal RNA is in ochre, proteins in blue. The active site is a small segment of rRNA, indicated in red.
Like DNA, most biologically active RNAs, including mRNA, tRNA, rRNA, snRNAs, and other non-coding RNAs, contain self-complementary sequences that allow parts of the RNA to fold[5] and pair with itself to form double helices. Analysis of these RNAs has revealed that they are highly structured. Unlike DNA, their structures do not consist of long double helices, but rather collections of short helices packed together into structures akin to proteins. In this fashion, RNAs can achieve chemical catalysis (like enzymes).[6] For instance, determination of the structure of the ribosome—an RNA-protein complex that catalyzes peptide bond formation—revealed that its active site is composed entirely of RNA.[7]
Structure
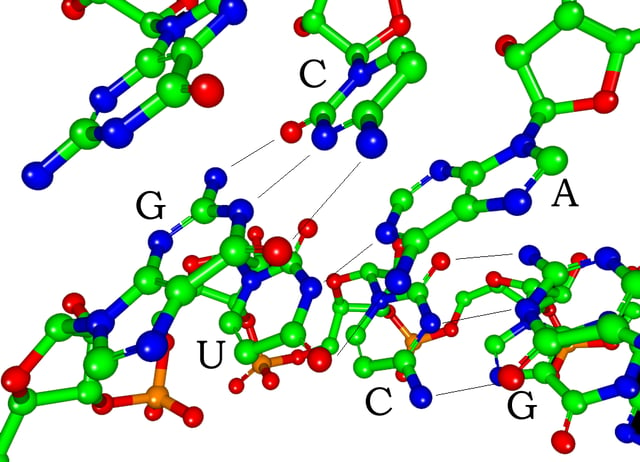
Watson-Crick base pairs in a siRNA (hydrogen atoms are not shown)
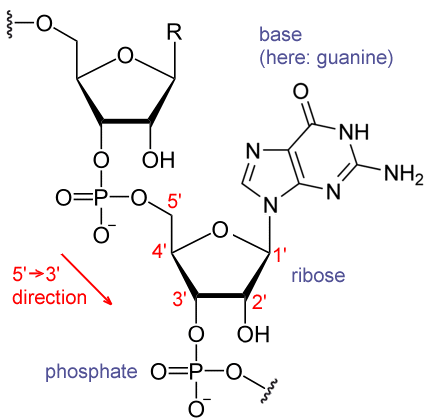
Chemical structure of RNA
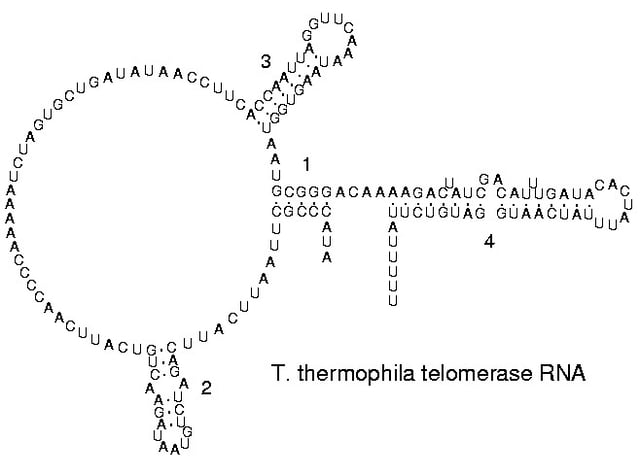
Secondary structure of a telomerase RNA.
Each nucleotide in RNA contains a ribose sugar, with carbons numbered 1' through 5'. A base is attached to the 1' position, in general, adenine (A), cytosine (C), guanine (G), or uracil (U). Adenine and guanine are purines, cytosine and uracil are pyrimidines. A phosphate group is attached to the 3' position of one ribose and the 5' position of the next. The phosphate groups have a negative charge each, making RNA a charged molecule (polyanion). The bases form hydrogen bonds between cytosine and guanine, between adenine and uracil and between guanine and uracil.[8] However, other interactions are possible, such as a group of adenine bases binding to each other in a bulge,[9] or the GNRA tetraloop that has a guanine–adenine base-pair.[8]
An important structural component of RNA that distinguishes it from DNA is the presence of a hydroxyl group at the 2' position of the ribose sugar. The presence of this functional group causes the helix to mostly take the A-form geometry,[10] although in single strand dinucleotide contexts, RNA can rarely also adopt the B-form most commonly observed in DNA.[11] The A-form geometry results in a very deep and narrow major groove and a shallow and wide minor groove.[12] A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is, not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone.[13]
RNA is transcribed with only four bases (adenine, cytosine, guanine and uracil),[14] but these bases and attached sugars can be modified in numerous ways as the RNAs mature. Pseudouridine (Ψ), in which the linkage between uracil and ribose is changed from a C–N bond to a C–C bond, and ribothymidine (T) are found in various places (the most notable ones being in the TΨC loop of tRNA).[15] Another notable modified base is hypoxanthine, a deaminated adenine base whose nucleoside is called inosine (I). Inosine plays a key role in the wobble hypothesis of the genetic code.[16]
There are more than 100 other naturally occurring modified nucleosides.[17] The greatest structural diversity of modifications can be found in tRNA,[18] while pseudouridine and nucleosides with 2'-O-methylribose often present in rRNA are the most common.[19] The specific roles of many of these modifications in RNA are not fully understood. However, it is notable that, in ribosomal RNA, many of the post-transcriptional modifications occur in highly functional regions, such as the peptidyl transferase center and the subunit interface, implying that they are important for normal function.[20]
The functional form of single-stranded RNA molecules, just like proteins, frequently requires a specific tertiary structure. The scaffold for this structure is provided by secondary structural elements that are hydrogen bonds within the molecule. This leads to several recognizable "domains" of secondary structure like hairpin loops, bulges, and internal loops.[21] Since RNA is charged, metal ions such as Mg2+ are needed to stabilise many secondary and tertiary structures.[22]
The naturally occurring enantiomer of RNA is D-RNA composed of D-ribonucleotides. All chirality centers are located in the D-ribose. By the use of L-ribose or rather L-ribonucleotides, L-RNA can be synthesized. L-RNA is much more stable against degradation by RNase.[23]
Like other structured biopolymers such as proteins, one can define topology of a folded RNA molecule. This is often done based on arrangement of intra-chain contacts within a folded RNA, termed as circuit topology.
Synthesis
Synthesis of RNA is usually catalyzed by an enzyme—RNA polymerase—using DNA as a template, a process known as transcription. Initiation of transcription begins with the binding of the enzyme to a promoter sequence in the DNA (usually found "upstream" of a gene). The DNA double helix is unwound by the helicase activity of the enzyme. The enzyme then progresses along the template strand in the 3’ to 5’ direction, synthesizing a complementary RNA molecule with elongation occurring in the 5’ to 3’ direction. The DNA sequence also dictates where termination of RNA synthesis will occur.[24]
Primary transcript RNAs are often modified by enzymes after transcription. For example, a poly(A) tail and a 5' cap are added to eukaryotic pre-mRNA and introns are removed by the spliceosome.
There are also a number of RNA-dependent RNA polymerases that use RNA as their template for synthesis of a new strand of RNA. For instance, a number of RNA viruses (such as poliovirus) use this type of enzyme to replicate their genetic material.[25] Also, RNA-dependent RNA polymerase is part of the RNA interference pathway in many organisms.[26]
Types of RNA
Overview
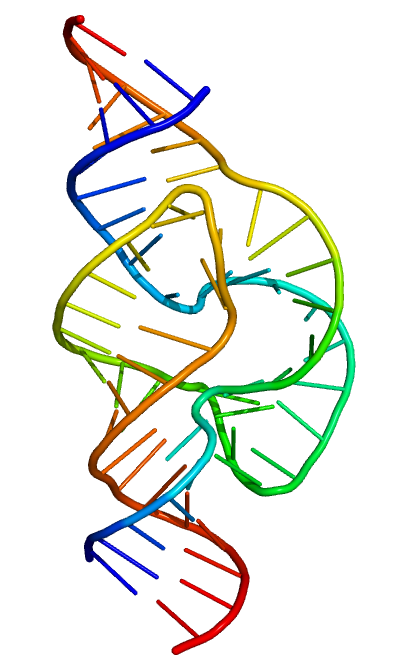
Structure of a hammerhead ribozyme, a ribozyme that cuts RNA
Messenger RNA (mRNA) is the RNA that carries information from DNA to the ribosome, the sites of protein synthesis (translation) in the cell. The coding sequence of the mRNA determines the amino acid sequence in the protein that is produced.[27] However, many RNAs do not code for protein (about 97% of the transcriptional output is non-protein-coding in eukaryotes[28][29][30][31]).
These so-called non-coding RNAs ("ncRNA") can be encoded by their own genes (RNA genes), but can also derive from mRNA introns.[32] The most prominent examples of non-coding RNAs are transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation.[4] There are also non-coding RNAs involved in gene regulation, RNA processing and other roles. Certain RNAs are able to catalyse chemical reactions such as cutting and ligating other RNA molecules,[33] and the catalysis of peptide bond formation in the ribosome;[7] these are known as ribozymes.
In length
According to the length of RNA chain, RNA includes small RNA and long RNA.[34] Usually, small RNAs are shorter than 200 nt in length, and long RNAs are greater than 200 nt long.[35] Long RNAs, also called large RNAs, mainly include long non-coding RNA (lncRNA) and mRNA. Small RNAs mainly include 5.8S ribosomal RNA (rRNA), 5S rRNA, transfer RNA (tRNA), microRNA (miRNA), small interfering RNA (siRNA), small nucleolar RNA (snoRNAs), Piwi-interacting RNA (piRNA), tRNA-derived small RNA (tsRNA)[36] and small rDNA-derived RNA (srRNA).[37]
In translation
Messenger RNA (mRNA) carries information about a protein sequence to the ribosomes, the protein synthesis factories in the cell. It is coded so that every three nucleotides (a codon) corresponds to one amino acid. In eukaryotic cells, once precursor mRNA (pre-mRNA) has been transcribed from DNA, it is processed to mature mRNA. This removes its introns—non-coding sections of the pre-mRNA. The mRNA is then exported from the nucleus to the cytoplasm, where it is bound to ribosomes and translated into its corresponding protein form with the help of tRNA. In prokaryotic cells, which do not have nucleus and cytoplasm compartments, mRNA can bind to ribosomes while it is being transcribed from DNA. After a certain amount of time, the message degrades into its component nucleotides with the assistance of ribonucleases.[27]
Transfer RNA (tRNA) is a small RNA chain of about 80 nucleotides that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation. It has sites for amino acid attachment and an anticodon region for codon recognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding.[32]
Ribosomal RNA (rRNA) is the catalytic component of the ribosomes. Eukaryotic ribosomes contain four different rRNA molecules: 18S, 5.8S, 28S and 5S rRNA. Three of the rRNA molecules are synthesized in the nucleolus, and one is synthesized elsewhere. In the cytoplasm, ribosomal RNA and protein combine to form a nucleoprotein called a ribosome. The ribosome binds mRNA and carries out protein synthesis. Several ribosomes may be attached to a single mRNA at any time.[27] Nearly all the RNA found in a typical eukaryotic cell is rRNA.
Regulatory RNA
The earliest known regulators of gene expression were proteins known as repressors and activators, regulators with specific short binding sites within enhancer regions near the genes to be regulated.[39] More recently, RNAs have been found to regulate genes as well. There are several kinds of RNA-dependent processes in eukaryotes regulating the expression of genes at various points, such as RNAi repressing genes post-transcriptionally, long non-coding RNAs shutting down blocks of chromatin epigenetically, and enhancer RNAs inducing increased gene expression.[40] In addition to these mechanisms in eukaryotes, both bacteria and archaea have been found to use regulatory RNAs extensively. Bacterial small RNA and the CRISPR system are examples of such prokaryotic regulatory RNA systems.[41] Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for discovering microRNAs (miRNAs), specific short RNA molecules that can base-pair with mRNAs.[42]
RNA interference by miRNAs
Post-transcriptional expression levels of many genes can be controlled by RNA interference, in which miRNAs, specific short RNA molecules, pair with meRNA regions and target them for degradation.[43] This antisense-based process involves steps that first process the RNA so that it can base-pair with a region of its target mRNAs. Once the base pairing occurs, other proteins direct the mRNA to be destroyed by nucleases.[40] Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for this discovery.[42]
Long non-coding RNAs
Next to be linked to regulation were Xist and other long noncoding RNAs associated with X chromosome inactivation. Their roles, at first mysterious, were shown by Jeannie T. Lee and others to be the silencing of blocks of chromatin via recruitment of Polycomb complex so that messenger RNA could not be transcribed from them.[44] Additional lncRNAs, currently defined as RNAs of more than 200 base pairs that do not appear to have coding potential,[45] have been found associated with regulation of stem cell pluripotency and cell division.[45]
Enhancer RNAs
The third major group of regulatory RNAs is called enhancer RNAs.[45] It is not clear at present whether they are a unique category of RNAs of various lengths or constitute a distinct subset of lncRNAs. In any case, they are transcribed from enhancers, which are known regulatory sites in the DNA near genes they regulate.[45][46] They up-regulate the transcription of the gene(s) under control of the enhancer from which they are transcribed.[45][47]
Regulatory RNA in prokaryotes
At first, regulatory RNA was thought to be a eukaryotic phenomenon, a part of the explanation for why so much more transcription in higher organisms was seen than had been predicted. But as soon as researchers began to look for possible RNA regulators in bacteria, they turned up there as well.[41] Currently, the ubiquitous nature of systems of RNA regulation of genes has been discussed as support for the RNA World theory.[40][48] Bacterial small RNAs generally act via antisense pairing with mRNA to down-regulate its translation, either by affecting stability or affecting cis-binding ability.[40] Riboswitches have also been discovered. They are cis-acting regulatory RNA sequences acting allosterically. They change shape when they bind metabolites so that they gain or lose the ability to bind chromatin to regulate expression of genes.[49][50]
In RNA processing

Uridine to pseudouridine is a common RNA modification.
Many RNAs are involved in modifying other RNAs. Introns are spliced out of pre-mRNA by spliceosomes, which contain several small nuclear RNAs (snRNA),[4] or the introns can be ribozymes that are spliced by themselves.[53] RNA can also be altered by having its nucleotides modified to nucleotides other than A, C, G and U. In eukaryotes, modifications of RNA nucleotides are in general directed by small nucleolar RNAs (snoRNA; 60–300 nt),[32] found in the nucleolus and cajal bodies. snoRNAs associate with enzymes and guide them to a spot on an RNA by basepairing to that RNA. These enzymes then perform the nucleotide modification. rRNAs and tRNAs are extensively modified, but snRNAs and mRNAs can also be the target of base modification.[54][55] RNA can also be methylated.[56][57]
RNA genomes
Like DNA, RNA can carry genetic information. RNA viruses have genomes composed of RNA that encodes a number of proteins. The viral genome is replicated by some of those proteins, while other proteins protect the genome as the virus particle moves to a new host cell. Viroids are another group of pathogens, but they consist only of RNA, do not encode any protein and are replicated by a host plant cell's polymerase.[58]
In reverse transcription
Reverse transcribing viruses replicate their genomes by reverse transcribing DNA copies from their RNA; these DNA copies are then transcribed to new RNA. Retrotransposons also spread by copying DNA and RNA from one another,[59] and telomerase contains an RNA that is used as template for building the ends of eukaryotic chromosomes.[60]
Double-stranded RNA
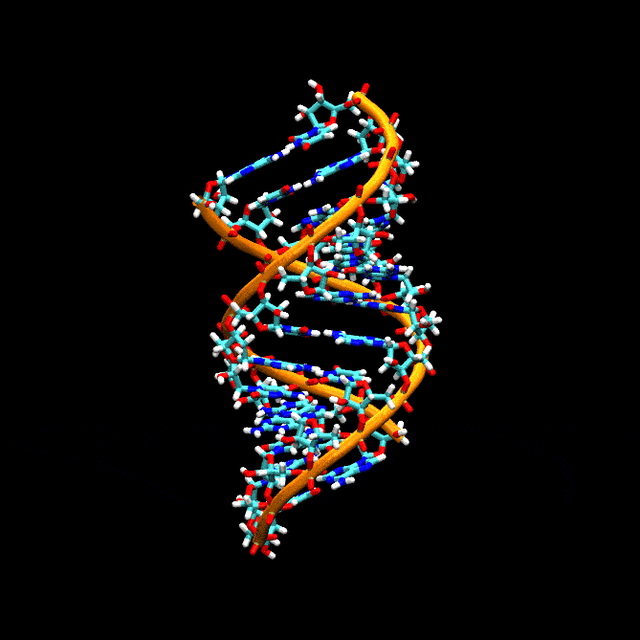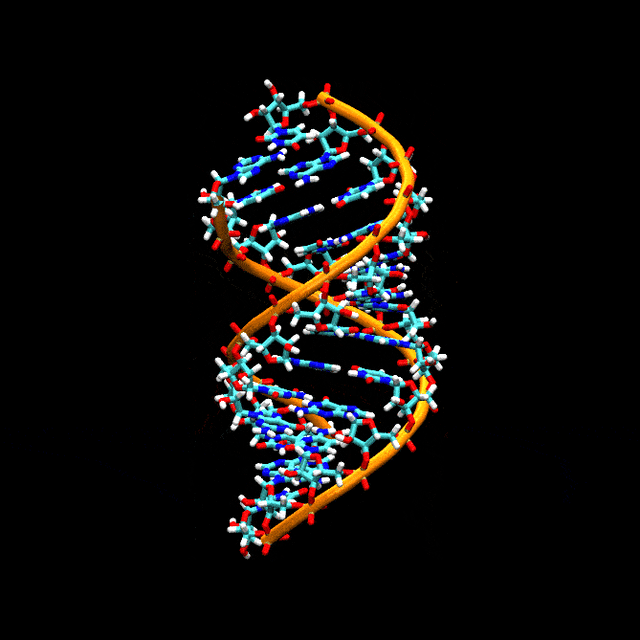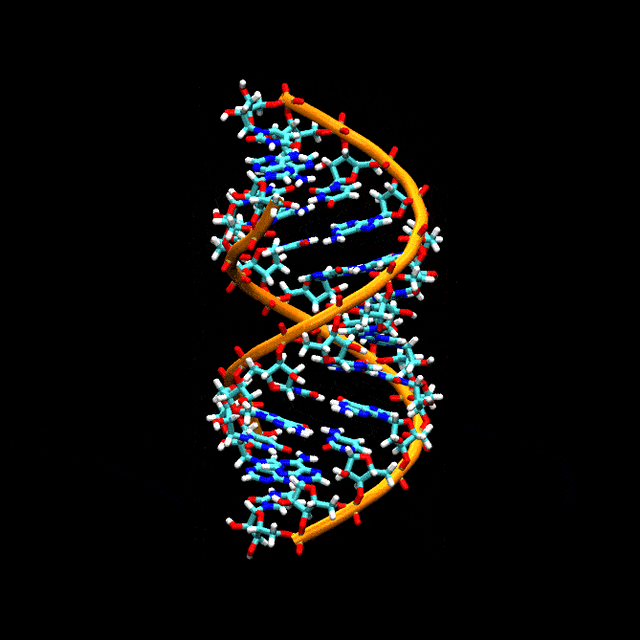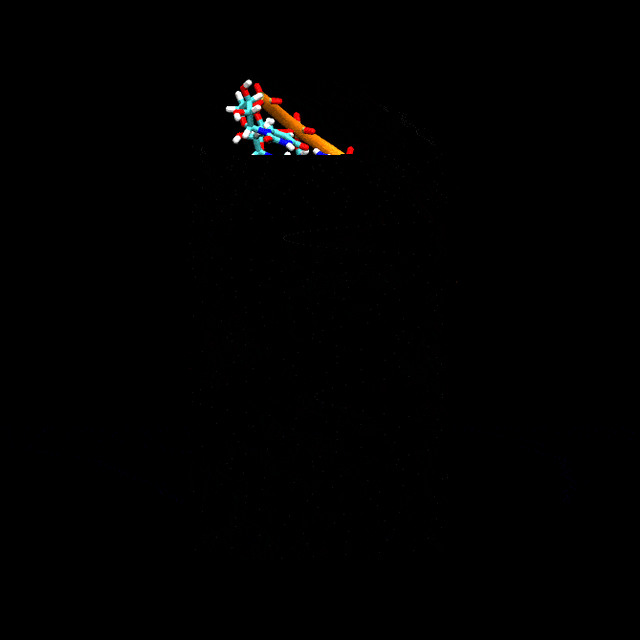
Double-stranded RNA
Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells, but with the replacement of thymine by uracil. dsRNA forms the genetic material of some viruses (double-stranded RNA viruses). Double-stranded RNA, such as viral RNA or siRNA, can trigger RNA interference in eukaryotes, as well as interferon response in vertebrates.[61][62][63][64]
Circular RNA
In the late 1970s, it was shown that there is a single stranded covalently closed, i.e. circular form of RNA expressed throughout the animal and plant kingdom (see circRNA).[65] circRNAs are thought to arise via a "back-splice" reaction where the spliceosome joins a downstream donor to an upstream acceptor splice site. So far the function of circRNAs is largely unknown, although for few examples a microRNA sponging activity has been demonstrated.
Key discoveries in RNA biology

Robert W. Holley, left, poses with his research team.
Research on RNA has led to many important biological discoveries and numerous Nobel Prizes. Nucleic acids were discovered in 1868 by Friedrich Miescher, who called the material 'nuclein' since it was found in the nucleus.[66] It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids. The role of RNA in protein synthesis was suspected already in 1939.[67] Severo Ochoa won the 1959 Nobel Prize in Medicine (shared with Arthur Kornberg) after he discovered an enzyme that can synthesize RNA in the laboratory.[68] However, the enzyme discovered by Ochoa (polynucleotide phosphorylase) was later shown to be responsible for RNA degradation, not RNA synthesis. In 1956 Alex Rich and David Davies hybridized two separate strands of RNA to form the first crystal of RNA whose structure could be determined by X-ray crystallography.[69]
The sequence of the 77 nucleotides of a yeast tRNA was found by Robert W. Holley in 1965,[70] winning Holley the 1968 Nobel Prize in Medicine (shared with Har Gobind Khorana and Marshall Nirenberg).
During the early 1970s, retroviruses and reverse transcriptase were discovered, showing for the first time that enzymes could copy RNA into DNA (the opposite of the usual route for transmission of genetic information). For this work, David Baltimore, Renato Dulbecco and Howard Temin were awarded a Nobel Prize in 1975. In 1976, Walter Fiers and his team determined the first complete nucleotide sequence of an RNA virus genome, that of bacteriophage MS2.[71]
In 1977, introns and RNA splicing were discovered in both mammalian viruses and in cellular genes, resulting in a 1993 Nobel to Philip Sharp and Richard Roberts. Catalytic RNA molecules (ribozymes) were discovered in the early 1980s, leading to a 1989 Nobel award to Thomas Cech and Sidney Altman. In 1990, it was found in Petunia that introduced genes can silence similar genes of the plant's own, now known to be a result of RNA interference.[72][73]
At about the same time, 22 nt long RNAs, now called microRNAs, were found to have a role in the development of C. elegans.[74] Studies on RNA interference gleaned a Nobel Prize for Andrew Fire and Craig Mello in 2006, and another Nobel was awarded for studies on the transcription of RNA to Roger Kornberg in the same year. The discovery of gene regulatory RNAs has led to attempts to develop drugs made of RNA, such as siRNA, to silence genes.[75] Adding to the Nobel prizes awarded for research on RNA in 2009 it was awarded for the elucidation of the atomic structure of the ribosome to Venki Ramakrishnan, Tom Steitz, and Ada Yonath.
Relevance for prebiotic chemistry and abiogenesis
In March 2015, complex DNA and RNA nucleotides, including uracil, cytosine and thymine, were reportedly formed in the laboratory under outer space conditions, using starter chemicals, such as pyrimidine, an organic compound commonly found in meteorites. Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), is one of the most carbon-rich compounds found in the Universe and may have been formed in red giants or in interstellar dust and gas clouds.[78]
See also
Biomolecular structure
DNA
History of RNA Biology
List of RNA Biologists
Macromolecule
RNA-based evolution
RNA origami
Transcriptome